Setting up Jupyter Lab integrated with Python, Julia, R on Windows Subsystem for Linux (Ubuntu)
Every guy gotta favourite OS. But, due to some constraints, all can't use that all the time. Being a Linux user, I had to shift to Windows 10 because of the barrier by MS Office and Dual booting often doesn't work. A few weeks before I learned about Windows Subsystem for Linux and my experience was great. If you are Linux pro stuck in Windows, this could be a relief. Basically, you can run a well-designed minimal version of Ubuntu inside Windows 10.
In this article, we will get familiar with,
>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>
* rpm: openssl-devel (Fedora, CentOS, RHEL)
* csw: libssl_dev (Solaris)
* brew: openssl@1.1 (Mac OSX)
If openssl is already installed, check that 'pkg-config' is in your
PATH and PKG_CONFIG_PATH contains a openssl.pc file. If pkg-config
is unavailable you can set INCLUDE_DIR and LIB_DIR manually via:
R CMD INSTALL --configure-vars='INCLUDE_DIR=... LIB_DIR=...'
-------------------------------------------------------------------
In this article, we will get familiar with,
- Activation of Windows Subsystem for Linux.
- Installation of Ubuntu (no GUI) using Microsoft App Store.
- Configuring Python3, Pip3 and Jupyter Lab.
- Installation of R in Ubuntu and integration of R with Jupyter Lab.
- Installation of Julia in Ubuntu along with Jupyter Lab integration.
- Accessing Windows files with Ubuntu.
Installation of Windows Subsystem for Linux
Windows don't ship with WSL by default, we need to install it via PowerShell.
Steps to follow,
1. Open PowerShell as Administrator and run following command.
"Enable-WindowsOptionalFeature -Online -FeatureName Microsoft-Windows-Subsystem-Linux"
2. Restart your computer after installation.
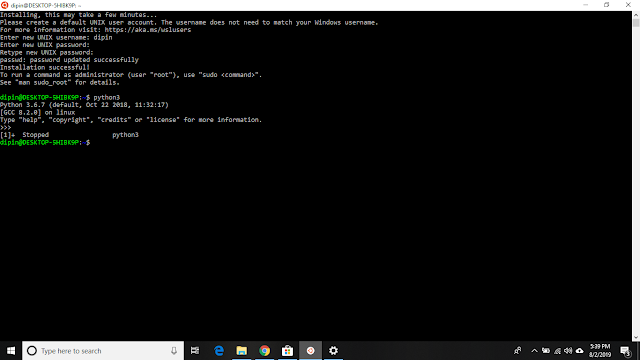
Installation of Ubuntu(minimal) from the Microsoft App Store.
Different Linux distros (like Ubuntu, Kali) are available in Microsoft Store. Right now we are going with Ubuntu.
1. Open the Microsoft Store and search for "Ubuntu".
2. Click on "Get", it will start downloading. After completion click on the "Launch" button and a bash equivalent will start running.
3. Let's do some initial set up, Enter a user name and password for Ubuntu.
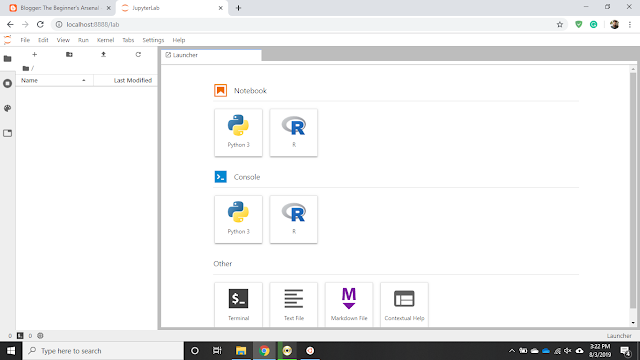
Configuring Python3, Pip3 and Jupyter Lab
1. Ubuntu on Windows got Python3 by default. Verify,
2. Upgrade Ubuntu to the latest. Shoot the commands,
"sudo apt update"
"sudo apt upgrade"
3. Time to install Pip3 from the repo, run the following in terminal,
"sudo apt install python3-pip"
4. Install Jupyter Lab using Pip3.
"sudo pip3 install jupyter-lab"
Restart Ubuntu and check if Jupyter Lab is running by typing "jupyter lab".
5. Copy and paste the displayed URL starting with "http://localhost:8888/lab" in a browser and check if Python is accessible.
The first part of our setup is over, let's proceed with R and Julia.
Installation of R in Ubuntu and integration of R with Jupyter Lab
Installing R and its libraries was a little tiring. I encountered with permission and dependency issues. So I'm suggesting to use R with sudo privileges and install some dependencies beforehand.
Few observed issues,
>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>
* rpm: openssl-devel (Fedora, CentOS, RHEL)
* csw: libssl_dev (Solaris)
* brew: openssl@1.1 (Mac OSX)
If openssl is already installed, check that 'pkg-config' is in your
PATH and PKG_CONFIG_PATH contains a openssl.pc file. If pkg-config
is unavailable you can set INCLUDE_DIR and LIB_DIR manually via:
R CMD INSTALL --configure-vars='INCLUDE_DIR=... LIB_DIR=...'
-------------------------------------------------------------------
ERROR: configuration failed for package ‘openssl’
* removing ‘/usr/local/lib/R/site-library/openssl’
* installing *source* package ‘xml2’ ...
** package ‘xml2’ successfully unpacked and MD5 sums checked
Package libxml-2.0 was not found in the pkg-config search path.
Perhaps you should add the directory containing `libxml-2.0.pc'
to the PKG_CONFIG_PATH environment variable
No package 'libxml-2.0' found
Package libxml-2.0 was not found in the pkg-config search path.
Perhaps you should add the directory containing `libxml-2.0.pc'
to the PKG_CONFIG_PATH environment variable
No package 'libxml-2.0' found
Using PKG_CFLAGS=
Using PKG_LIBS=-lxml2
------------------------- ANTICONF ERROR ---------------------------
Configuration failed because libxml-2.0 was not found. Try installing:
* deb: libxml2-dev (Debian, Ubuntu, etc)
* rpm: libxml2-devel (Fedora, CentOS, RHEL)
* csw: libxml2_dev (Solaris)
If libxml-2.0 is already installed, check that 'pkg-config' is in your
PATH and PKG_CONFIG_PATH contains a libxml-2.0.pc file. If pkg-config
is unavailable you can set INCLUDE_DIR and LIB_DIR manually via:
R CMD INSTALL --configure-vars='INCLUDE_DIR=... LIB_DIR=...'
--------------------------------------------------------------------
------------------------- ANTICONF ERROR ---------------------------
Configuration failed because openssl was not found. Try installing:
* deb: libssl-dev (Debian, Ubuntu, etc)
* rpm: openssl-devel (Fedora, CentOS, RHEL)
* csw: libssl_dev (Solaris)
* brew: openssl@1.1 (Mac OSX)
If openssl is already installed, check that 'pkg-config' is in your
PATH and PKG_CONFIG_PATH contains a openssl.pc file. If pkg-config
is unavailable you can set INCLUDE_DIR and LIB_DIR manually via:
R CMD INSTALL --configure-vars='INCLUDE_DIR=... LIB_DIR=...'
-------------------------------------------------------------------
ERROR: configuration failed for package ‘openssl’
* removing ‘/usr/local/lib/R/site-library/openssl’
* installing *source* package ‘xml2’ ...
** package ‘xml2’ successfully unpacked and MD5 sums checked
Package libxml-2.0 was not found in the pkg-config search path.
Perhaps you should add the directory containing `libxml-2.0.pc'
to the PKG_CONFIG_PATH environment variable
No package 'libxml-2.0' found
Package libxml-2.0 was not found in the pkg-config search path.
Perhaps you should add the directory containing `libxml-2.0.pc'
to the PKG_CONFIG_PATH environment variable
No package 'libxml-2.0' found
Using PKG_CFLAGS=
Using PKG_LIBS=-lxml2
------------------------- ANTICONF ERROR ---------------------------
Configuration failed because libxml-2.0 was not found. Try installing:
* deb: libxml2-dev (Debian, Ubuntu, etc)
* rpm: libxml2-devel (Fedora, CentOS, RHEL)
* csw: libxml2_dev (Solaris)
If libxml-2.0 is already installed, check that 'pkg-config' is in your
PATH and PKG_CONFIG_PATH contains a libxml-2.0.pc file. If pkg-config
is unavailable you can set INCLUDE_DIR and LIB_DIR manually via:
R CMD INSTALL --configure-vars='INCLUDE_DIR=... LIB_DIR=...'
--------------------------------------------------------------------
------------------------- ANTICONF ERROR ---------------------------
Configuration failed because libcurl was not found. Try installing:
* deb: libcurl4-openssl-dev (Debian, Ubuntu, etc)
* rpm: libcurl-devel (Fedora, CentOS, RHEL)
* csw: libcurl_dev (Solaris)
If libcurl is already installed, check that 'pkg-config' is in your
PATH and PKG_CONFIG_PATH contains a libcurl.pc file. If pkg-config
is unavailable you can set INCLUDE_DIR and LIB_DIR manually via:
R CMD INSTALL --configure-vars='INCLUDE_DIR=... LIB_DIR=...'
* removing ‘/usr/local/lib/R/site-library/openssl’
* installing *source* package ‘xml2’ ...
** package ‘xml2’ successfully unpacked and MD5 sums checked
Package libxml-2.0 was not found in the pkg-config search path.
Perhaps you should add the directory containing `libxml-2.0.pc'
to the PKG_CONFIG_PATH environment variable
No package 'libxml-2.0' found
Package libxml-2.0 was not found in the pkg-config search path.
Perhaps you should add the directory containing `libxml-2.0.pc'
to the PKG_CONFIG_PATH environment variable
No package 'libxml-2.0' found
Using PKG_CFLAGS=
Using PKG_LIBS=-lxml2
------------------------- ANTICONF ERROR ---------------------------
Configuration failed because libxml-2.0 was not found. Try installing:
* deb: libxml2-dev (Debian, Ubuntu, etc)
* rpm: libxml2-devel (Fedora, CentOS, RHEL)
* csw: libxml2_dev (Solaris)
If libxml-2.0 is already installed, check that 'pkg-config' is in your
PATH and PKG_CONFIG_PATH contains a libxml-2.0.pc file. If pkg-config
is unavailable you can set INCLUDE_DIR and LIB_DIR manually via:
R CMD INSTALL --configure-vars='INCLUDE_DIR=... LIB_DIR=...'
--------------------------------------------------------------------
------------------------- ANTICONF ERROR ---------------------------
Configuration failed because openssl was not found. Try installing:
* deb: libssl-dev (Debian, Ubuntu, etc)
* rpm: openssl-devel (Fedora, CentOS, RHEL)
* csw: libssl_dev (Solaris)
* brew: openssl@1.1 (Mac OSX)
If openssl is already installed, check that 'pkg-config' is in your
PATH and PKG_CONFIG_PATH contains a openssl.pc file. If pkg-config
is unavailable you can set INCLUDE_DIR and LIB_DIR manually via:
R CMD INSTALL --configure-vars='INCLUDE_DIR=... LIB_DIR=...'
-------------------------------------------------------------------
ERROR: configuration failed for package ‘openssl’
* removing ‘/usr/local/lib/R/site-library/openssl’
* installing *source* package ‘xml2’ ...
** package ‘xml2’ successfully unpacked and MD5 sums checked
Package libxml-2.0 was not found in the pkg-config search path.
Perhaps you should add the directory containing `libxml-2.0.pc'
to the PKG_CONFIG_PATH environment variable
No package 'libxml-2.0' found
Package libxml-2.0 was not found in the pkg-config search path.
Perhaps you should add the directory containing `libxml-2.0.pc'
to the PKG_CONFIG_PATH environment variable
No package 'libxml-2.0' found
Using PKG_CFLAGS=
Using PKG_LIBS=-lxml2
------------------------- ANTICONF ERROR ---------------------------
Configuration failed because libxml-2.0 was not found. Try installing:
* deb: libxml2-dev (Debian, Ubuntu, etc)
* rpm: libxml2-devel (Fedora, CentOS, RHEL)
* csw: libxml2_dev (Solaris)
If libxml-2.0 is already installed, check that 'pkg-config' is in your
PATH and PKG_CONFIG_PATH contains a libxml-2.0.pc file. If pkg-config
is unavailable you can set INCLUDE_DIR and LIB_DIR manually via:
R CMD INSTALL --configure-vars='INCLUDE_DIR=... LIB_DIR=...'
--------------------------------------------------------------------
------------------------- ANTICONF ERROR ---------------------------
Configuration failed because libcurl was not found. Try installing:
* deb: libcurl4-openssl-dev (Debian, Ubuntu, etc)
* rpm: libcurl-devel (Fedora, CentOS, RHEL)
* csw: libcurl_dev (Solaris)
If libcurl is already installed, check that 'pkg-config' is in your
PATH and PKG_CONFIG_PATH contains a libcurl.pc file. If pkg-config
is unavailable you can set INCLUDE_DIR and LIB_DIR manually via:
R CMD INSTALL --configure-vars='INCLUDE_DIR=... LIB_DIR=...'
--------------------------------------------------------------------
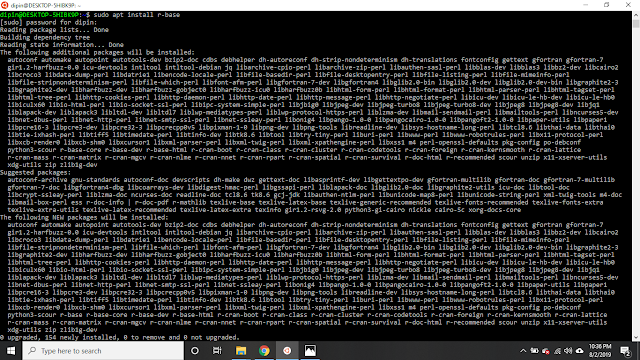
1. Let's start with installing R, open Ubuntu and run specific apt command.
2. Check if R is installed correctly, press R in the terminal and see,
1. Let's start with installing R, open Ubuntu and run specific apt command.
"sudo apt install r-base"
For some users, this won't work. In the case, please do an "apt update" and retry. Or for the official documentation - https://cran.r-project.org/
2. Check if R is installed correctly, press R in the terminal and see,
3. Before jumping into Jupyter Lab integration, install few dependencies that are needed.
"sudo apt install libssl-dev libxml2-dev libcurl4-openssl-dev"
4. Now, let's integrate R to Jupyter Lab, do the following steps sequentially,
>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>
Error in IRkernel::installspec() :
jupyter-client has to be installed but “jupyter kernelspec --version” exited with code 127.
> The issue occurs when jupyter lab doesn't got sudo privilege but R has or vice versa.
>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>
5. Check if R is integrated to Jupyter Lab, Restart Ubuntu and run "'jupyter lab"
The R part is over, now make Julia work,
Installation of Julia in Ubuntu along with Jupyter Lab integration
1. Go to https://julialang.org/downloads/, and copy the target link for Linux Binary. Use wget to download using Ubuntu terminal.
- install.packages("devtools")
- devtools::install_github("IRkernel/IRkernel")
- IRkernel::installspec()
If you are getting any lock error(happens when installation got aborted) try with following params,
"install.packages("devtools", dependencies=TRUE, INSTALL_opts = c('--no-lock')) "
>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>
Error in IRkernel::installspec() :
jupyter-client has to be installed but “jupyter kernelspec --version” exited with code 127.
> The issue occurs when jupyter lab doesn't got sudo privilege but R has or vice versa.
>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>
5. Check if R is integrated to Jupyter Lab, Restart Ubuntu and run "'jupyter lab"
The R part is over, now make Julia work,
Installation of Julia in Ubuntu along with Jupyter Lab integration
1. Go to https://julialang.org/downloads/, and copy the target link for Linux Binary. Use wget to download using Ubuntu terminal.
"wget https://julialang-s3.julialang.org/bin/linux/x64/1.1/julia-1.1.1-linux-x86_64.tar.gz"
2. Extract the zip file and create a soft link of "bin/julia" to "/usr/local/bin/julia". Please refer the attached screen shot or refer to this link - "https://stackoverflow.com/questions/41815059/installing-julia-v0-5-on-ubuntu-16-04-while-v0-6-is-installed"
Check if the installation is correct.
Check if the installation is correct.
3. Let's integrate Julia to Jupyter Lab. The best resource I found is - https://datatofish.com/add-julia-to-jupyter/.
Get into Julia shell, and do these sequentially,
Get into Julia shell, and do these sequentially,
- using Pkg
- Pkg.add("IJulia")
4. Let's see if Jupyter Lab can detect Julia, Restart Ubuntu and type "jupyter lab".
So, that's it. I hope everything is up. If you're stuck in any step, please comment below.
One more tip, which I found quite useful.
> In Ubuntu, the Windows partitions are available in /mnt/ directory. You can directly work on those files and duplication isn't needed. You may also consider adding you windows workspace folder to bashrc.
One more tip, which I found quite useful.
> In Ubuntu, the Windows partitions are available in /mnt/ directory. You can directly work on those files and duplication isn't needed. You may also consider adding you windows workspace folder to bashrc.















Comments
Post a Comment